Galaxy Usage on HPCC
9 minute read
What is Galaxy?
Galaxy is a free and open source scientific platform designed to make research accessible and reproducible. The HPCC has configured Galaxy to work on the cluster as an OnDemand application, allowing users to launch and maintain their very own Galaxy platform. The following sections below will explain how to use Galaxy on the cluster.
How to start a Galaxy session?
To start a Galaxy session, you must access our OnDemand instance and log in. You can find more information on how to access our OnDemand instance here. Once you are logged in, select the “Galaxy” application from the “Interactive Apps” tab as you would any other OnDemand application. Once selected, you need to fill out the following fields below:
Database Directory: This field is specific to Galaxy, when galaxy is launched a ‘.galaxy’ directory needs to be created. This directory will contain all files related to your Galaxy instance such as configuration files, databases, tool files, etc. Here, you would specify where you would want the ‘.galaxy’ to reside. If no path is given then, by default the directory will be created in your “rhome” directory.
Number of cores: This field specifies the number of CPU cores you would like to allocate to your Galaxy session. The amount allocated will be used for all local running jobs. The default amount of cores allocated is 4.
Memory in GB: Similar to the “Number of cores” field, this entry specifies the amount of memory you would like to allocate to your Galaxy session. The amount allocated will be used for all local running jobs. The default amount of memory allocated is 6 GBs.
Job runtime: This field specifies the amount of time your Galaxy session will be active. The Maximum runtime is determined by the HPCC’s queue policies, which can be found here
Partition: This field specifies the partition which your Galaxy session will be dispatched to. Depending on which partition is specified, the maximum runtime may vary. Please refer to the HPCC’s queue policies for more information.
Additional Slurm Arguments: This field specifies any additional slurm flags. If a GPU node is requested then the “–gres” flag will need to be added.
Galaxy tool runner: This field is specific to Galaxy, it specifies which “runner” to use for the Galaxy session. A “runner” is how jobs will be ran during your Galaxy session. If “local” is chosen then all jobs ran from within Galaxy, will be local to the machine from which Galaxy is running on. If the session is terminated, then so will all active running Galaxy jobs. If “cluster” is chosen then all jobs will be submitted via slurm. This includes workflows, file uploads, and tools. If the Galaxy session is terminated then all jobs will still continue to run until they are completed. Please refer to the following section for more information on how the “cluster” job runner works.
After you have filled on the fields, click on the “Launch” button to have your Galaxy session queued up to run.
Galaxy on OnDemand
Once your Galaxy session is running, a user is automatically created for you to access your Galaxy instance with. This user has admin privileges within your Galaxy instance. This allows for you to have more control when it comes to using Galaxy via OnDemand on the cluster. With a Galaxy admin account you are able to do the following:
- Install tools from Galaxy’s tool shed repository, which can include tools not installed on the cluster.
- Upload files locally from your computer and from the cluster directly into Galaxy.
- Submit jobs to slurm from within your Galaxy session.
- Execute Galaxy specific admin commands and or features.
As mentioned previously, a ‘.galaxy’ directory is created when a Galaxy session is launched for the first time. This directory is where all of your specific Galaxy files and dependencies are stored, such as tools installed via the tool shed, environment module tool files, other configuration files, and Galaxy’s database. You are able to freely modify any file within this directory and should be able to see any changes made once Galaxy is reloaded. If you delete this directory, then the directory will be re-made entirely new next time you launch Galaxy.
The HPCC admins have also added some extra features to Galaxy that allow users to interact with the cluster from within Galaxy. The next section will go into detail on these features and how they work.
Extra Galaxy Features
When starting a Galaxy session, you have the option to select which ‘runner’ to use for your Galaxy session. If the runner ‘cluster’ is chosen then all jobs started within Galaxy will be submitted to slurm. When selecting this runner, you will see a cog icon added to the menu bar for your Galaxy session. Clicking this icon will create a drop-down menu where you can configure the parameters used for all your jobs submitted via slurm from within Galaxy. The current default parameters are 2 CPU cores and 2 GBs of memory. The parameters set will be saved within your Galaxy database and will carry over into your next Galaxy session. Below you can see an example on how to modify the parameters for jobs submitted through Galaxy.

The cluster’s environment module system has been configured to work with Galaxy via Galaxy’s tool configuration file system. You can interact with any module on the cluster through Galaxy via the ‘Environment modules’ section in Galaxy’s Tool bar. Below you can see where this section is located.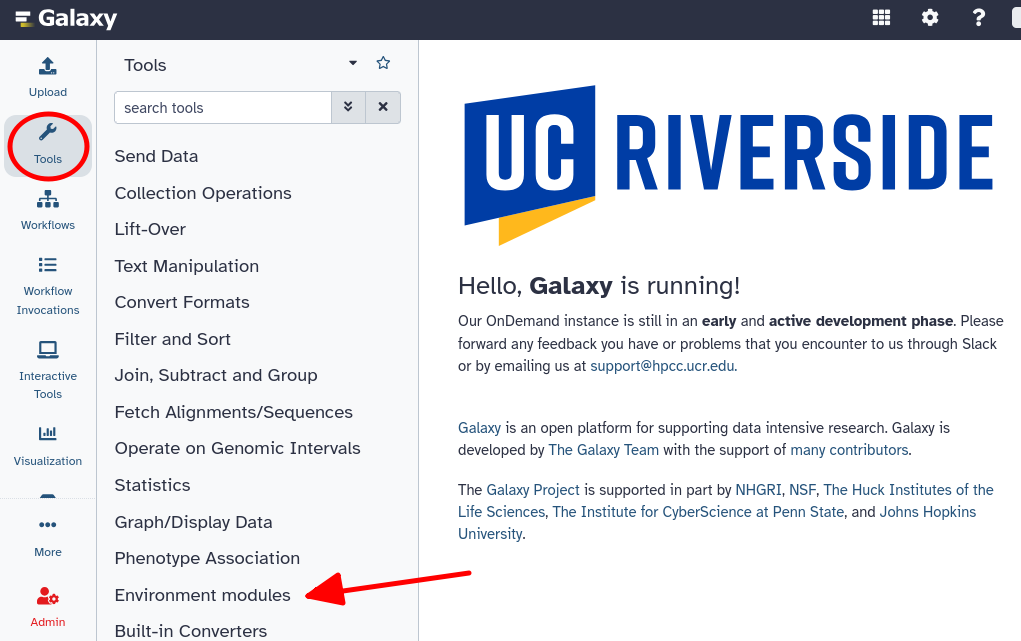
When interacting with an environment module tool in Galaxy, you will notice that each tool follows the same format. This is because since each environment module is unique, trying to create a unique configuration file for each module would be time-consuming. Instead, all environment module tools are created with a template that allows each tool to still be run from within Galaxy, using their command line specific commands. If this setup isn’t to your liking, you can either install the tool directly from Galaxy’s tool shed repository or edit the specific environment module tool configuration file. All environment module tool configurations can be found under your ‘.galaxy’ directory in the ‘tools’ directory under ‘modules’. Below is an example image of how an environment module tool looks, the tool shown is fastqc.
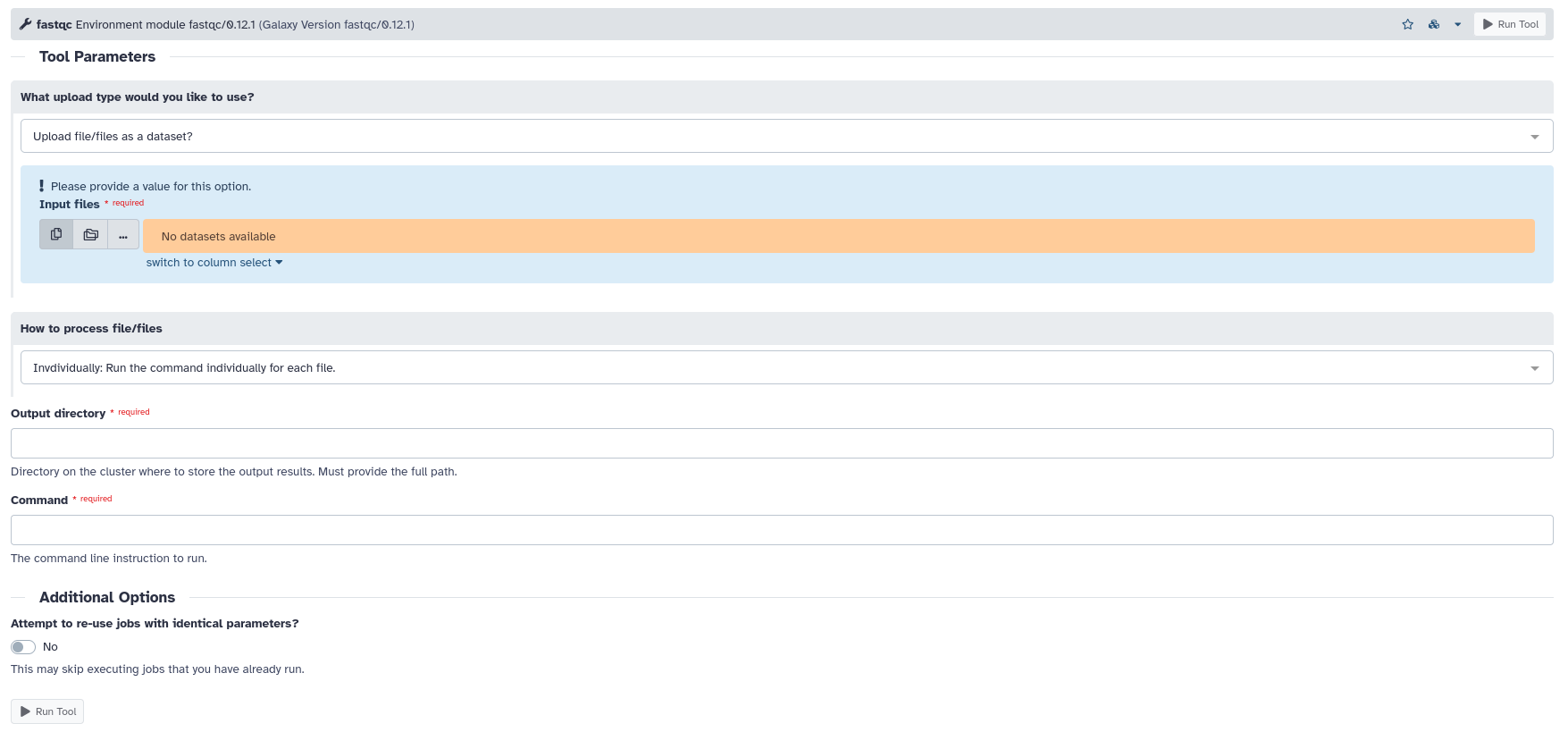
An explanation on what each parameter does can be found under each environment module tool. This manual will briefly explain what each parameter does, using the fastqc tool as an example.
What upload type would you like to use: This field specifies the files you would like to be used as input for the tool. Here you can either provide a ‘Dataset’ which is Galaxy’s custom format for files, or you can provide the full path of a file on the cluster as an input. One benefit for this method is that you don’t need to directly upload a file to the Galaxy in order for it to be used as input.
How to process file or files: This field specifies how you want the input files to be processed. Here you can either choose to process files individually or grouped. If you chose to process files individually, then the given command will run on all input files individually. Below you can see an example of how this would look using the fastqc tool via the command line.
fastqc -i file1;
fastqc -i file2;
fastqc -i file3;
...
If you chose to process files in a group, then all the files will be used as a single input to the command. Below you can see an example of how this would look using the fastqc tool via the command line.
fastqc -i file1 file2 file3 ...
Output directory: This field specifies the path on the cluster to the directory where output files will be dumped into. Each tool also has logic to parse a given output path and upload each output file automatically to Galaxy as a Dataset file.
Command: This field specifies the command the tool will use when it is executing. When entering the command, any inputs or outputs need to be specified as ‘$input’ or ‘$output’. This is because HPCC admins have configured the tool to replace these variables with the given input files and the path to the desired output directory. Below you can see an example of how this would look like, again using the fastqc tool via the command line.
fastqc -i $input -o $output
If a tool is installed from Galaxy’s tool shed repository and the same environment module tool corresponds to the specific installed tool, then the tool installed via Galaxy’s tool shed repository will take precedence over the environment module and act as the default. Most tools from Galaxy’s tool shed repository are unique to the tool they were made for, so they will have custom parameters that reflect this. This guide will not go over how to modify a tool configuration file, instead please refer to the Galaxy’s documentation on the topic.
Common Issues
While the HPCC admins have configured Galaxy on the cluster to be in a usable state, due to how detailed and large the Galaxy project is—there are still some issues the HPCC admins are still actively working to resolve. The following section details these issues. It is important to note that these issues do not hinder the usability of Galaxy on the cluster.
LocalProtocolError
When a user uploads a file from their local computer to their running Galaxy session, the following debug message can be seen in the ‘output.log’ file created for every OnDemand session.
h11._util.LocalProtocolError: Too much data for declared Content-Length
The local file will still be uploaded successfully to a user’s Galaxy session and can still be used normally within Galaxy.
Proxy Error 502
When a user attempts to install tools via Galaxy’s tool shed repository or tries to resolve dependencies for Galaxy specific tools, the following message will be displayed after 3 to 5 minutes.

The ‘output.log’ file shows no indication of this error, and while the message is still displayed—the task will still run in the background until it is fully complete. Meaning that tools installed via Galaxy’s tool shed repository will install successfully and dependencies will still be resolved after awhile. The progress of such tasks can be viewed in the ‘output.log’ file for a user’s OnDemand session.
This section will be updated continuously as issues are resolved or encountered. If you encounter any issues while using Galaxy on the cluster, please report them to our support email support@hpcc.ucr.edu